Automating away the "elbow method"
2018-09-24
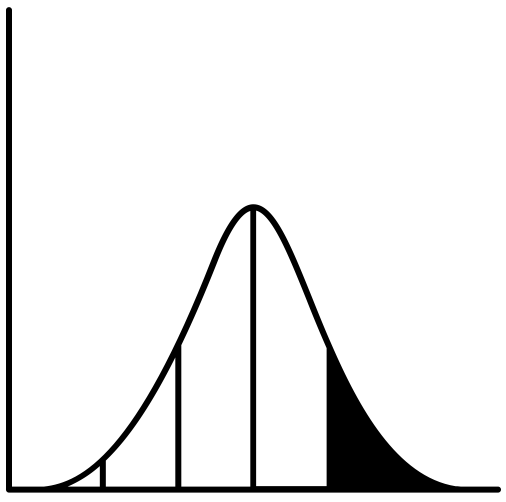
For some types of unsupervised learning analyses, machine learning practitioners have typically needed to examine a plot and make a somewhat subjective judgement call to tune the model (the so-called "elbow method"). I can think of two examples of this but others certainly exist:
1) In any sort of clustering analysis: finding the appropriate number of clusters by plotting the within-cluster sum of squares against the number of clusters.
2) When reducing feature space via PCA or a Factor Analysis: using a Scree plot to determine the number of components/factors to extract.
For one-off analyses, using your eyeballs and some subjectivity might be fine, but what if you are using these methods as part of a pipeline in an automated process? I came across a very simple and elegant solution to this, which is described by Mu Zhu in this paper. Lots of heuristics exist to solve this but I've found this method to be particularly robust.
Zhu's idea is to generate the data you would typically generate to identify the elbow/kink. Then, he treats this data as a composite of two different samples, separated by the cutoff he is trying to identify. He loops through all possible cutoffs, in an attempt to find the cutoff that maximizes the profile log-likelihood (using sample means and a pooled SD in the calculations). Here's my stab at implementing Zhu's method:
""" Automagical elbow finder """ import numpy as np from scipy.stats import norm def find_optimal_k(data): """ Provide a numpy array, returns index of that array to serve as elbow, cut-off point """ def __calc_logl(series, mu, sd): """ Helper function to calculate log-likelihood """ logl = 0 for i in series: logl += np.log(norm.pdf(i, mu, sd)) return logl profile_logl = [] # Loop through all possible pairs of series within the array for q in range(1, len(data)): n = len(data) s1 = data[0:q] s2 = data[q:] # Calculate means and standard deviations of both series mu1 = s1.mean() mu2 = s2.mean() sd1 = s1.std() sd2 = s2.std() sd_pooled = np.sqrt((((q-1)*(sd1**2)+(n-q-1)*(sd2**2)) / (n-2))) # Calculate profile log-likelihood profile_logl.append(calc_logl(s1, mu1, sd_pooled) + calc_logl(s2, mu2, sd_pooled)) return np.argmax(profile_logl) + 1
© Roni Kobrosly 2022